AI based prediction of adverse drug reactions
The post-marketing reports of adverse events for approved drugs and their combinations are available in VigiBase, FAERS, VAERS and JADER databases. However, some approved drugs and combinations are still missing there. Therefore, we have started developing an AI-based models to predict adverse events for the missing drugs and combinations in this project.
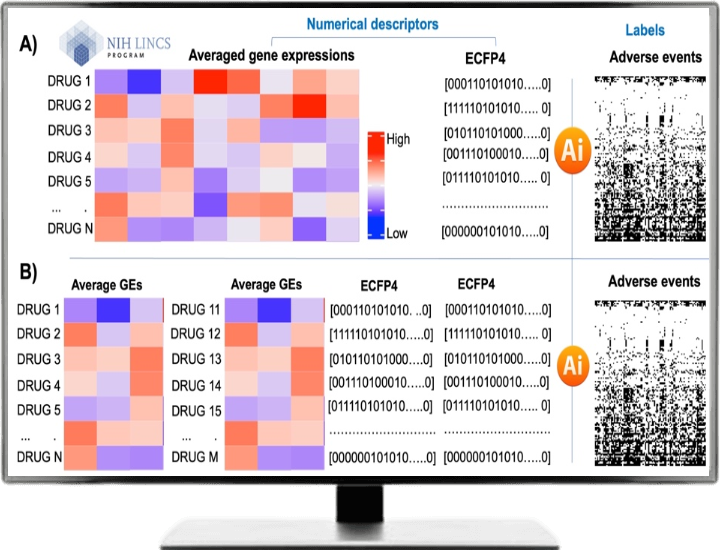
AI models are trained based on the drug’s structural fingerprints (ECFP4) and transcriptomic profiles from the LINCS L1000 database (https://lincsproject.org/LINCS/). For the sake of simplicity, we take an average of gene expression values across all cell lines and time points, resulting in a 12,328 x 3492 matrix (12,328 genes and 3492 drugs). ECFP4 has a bit string of length 2048.
We combine both vectors so that each drug will be represented by a descriptor vector of length 14376 (12,328+ 2048). For the drug combination model, the length of the descriptor vector is doubled. Integrating transcriptomic data from the LINCS L1000 dataset with chemical structural fingerprints enabled us to capture molecular and structural information, providing a more comprehensive representation of the compounds for adverse events prediction. Following figure shows the workflow of our proposed method.
