- info@bioicawtech.com
- Helsinki, Finland
- Request a quote
Pharamocofiler
-
BioICAWTech > Pharamocofiler
Pharamocofiler
Hamonizes and provide visual analytics on drug response data. Portal also integrates ML based live prediction of drug responses for uploaded any compounds.
Single-targeted drugs have demonstrated clinical success, it is unequivocal that single-targeted drugs often exhibit limited efficacy against complex diseases, such as cancers, whose development and treatment is dependent on several biological processes occurring simultaneously.
A comprehensive understanding of primary, secondary and inactive targets becomes essential in the quest for effective treatments for cancer and other indications.
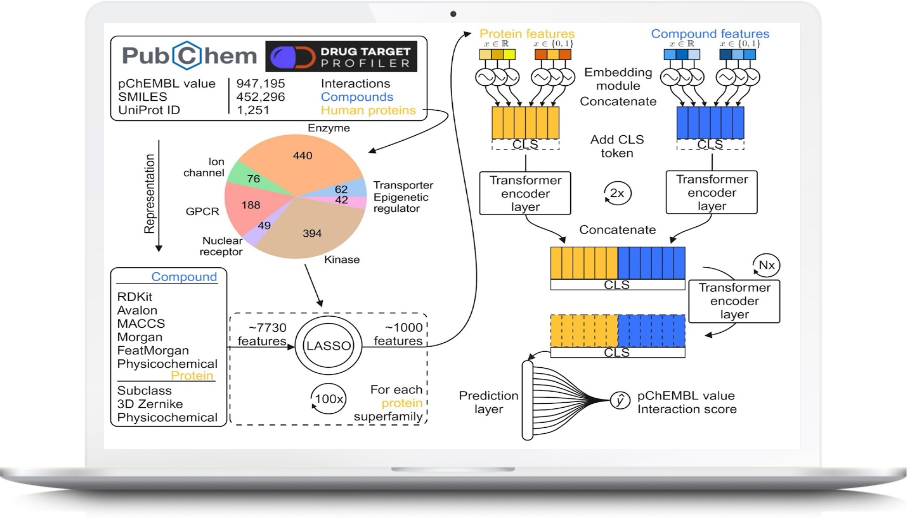
Most FDA-approved drugs bind with only a small fraction of potent targets with average number of potent targets per approved drugs < 7 as reported in one of our scientist’s study (MICHA reference).
Therefore, our team members have recently developed AI based prediction method that is able to accurately identify any drugs/compounds for >1500 human protein targets across seven super families i.e. GPCRs, Kinase, Enzymes, Nuclear receptors, Ion channels, epigenetic regulators, and transporters. Nine different descriptor sets were meticulously examined to identify optimal signature descriptors for predicting novel drug-target binding affinities. Framework for our AI based prediction method across seven superfamilies is shown in the following figure.
Our proposed method has been rigorously tested on 0.2M endpoints from independent dataset and outperformed all state-of-art prediction methods for drug-target affinity prediction. Manuscript is currently review.
We are now further improving the method and extending the method to six additional superfamilies namely: Adhesion, Membrane receptors, secreted proteins, structural proteins, Transcription factors and Surface antigen. As per our information there is no such comprehensive drug-target prediction method that can systemically screen millions of preclinical compounds and approved drugs across 13 superfamilies of protein targets.
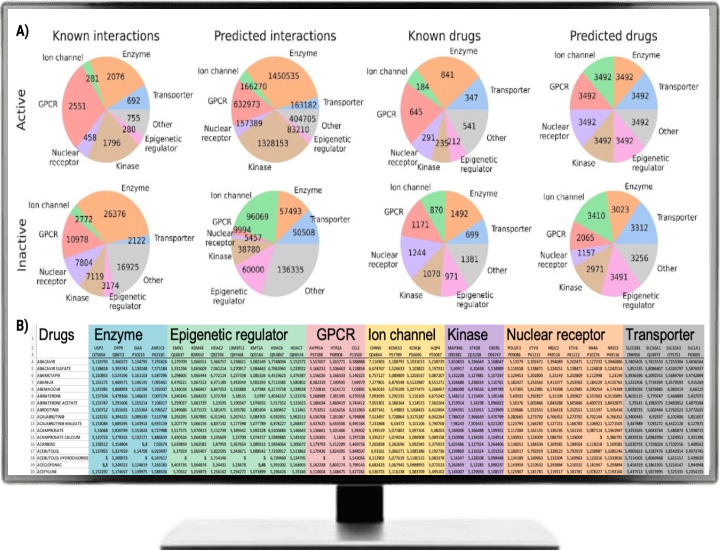
There are 3492 approved drugs (including salts) in ChEMBL33. But only a handful of approved drugs are experimentally tested against >50 human targets. Therefore, after successfully, testing and deploying drug-target based method, we went one step ahead and tried to complete drug-target profiles at full proteome (for 1251 human proteins). Figure 2A shows the pie charts for experimentally tested and predicted drug targets across seven superfamilies, whereas Figure 2B shows complete drug-target matrix for 3492 approved drugs (opening new avenues of drug repurposing).
